逐時變化與填入nc檔案
Table of contents
背景
- 環保署全年排放量乘上月變化係數
- 排放量整體處理原則參見處理程序總綱、針對植物源之處理及龐大
.dbf檔案之讀取,為此處之前處理。
程式分段說明
初始段
- 引用模組。此處用到include3.py的dt2jul, jul2dt, disc
kuang@node03 /nas1/TEDS/teds11/biog
$ cat -n bioginc.py
1 """usage:
2 python MM(month in 2 digits)
3 1.Domain is determined by the template chosen.
4 2.One file(month) at a time. The resultant file will not be overwritten.
5 3.nc files may be corrupted if not properly written. must be remove before redoing.
6 """
7 import numpy as np
8 from PseudoNetCDF.camxfiles.uamiv.Memmap import uamiv
9 from PseudoNetCDF.pncgen import pncgen
10 from pandas import *
11 from calendar import monthrange
12 import sys, os, subprocess
13 import netCDF4
14 import twd97
15 import datetime
16 from include3 import dt2jul, jul2dt, disc
17
18
19
- 讀取全年逐月排放檔
20 #Main
21 #import the gridded area sources
22 P='/nas1/TEDS/teds11/biog/'
23 fname=P+'biogrid2019.csv'
24 df = read_csv(fname)
25
- 讀入引數(年月共4碼)、計算起迄日期
26 #time and space initiates
27 mm=sys.argv[1]
28 mo=int(mm)
29 yr=2019
30 ntm=(monthrange(yr,mo)[1]+2)*24+1
31 bdate=datetime.datetime(yr,mo,1)+datetime.timedelta(days=-1+8./24)
32 edate=bdate+datetime.timedelta(days=monthrange(yr,mo)[1]+3)
33
nc模版的準備
34 #prepare the uamiv template
35 fname='fortBE.413_teds10.biog'+mm+'.nc'
36 try:
37 nc = netCDF4.Dataset(fname, 'r+')
38 except:
39 os.system('cp '+P+'template_d4.nc '+fname)
40 nc = netCDF4.Dataset(fname, 'r+')
41 V=[list(filter(lambda x:nc.variables[x].ndim==j, [i for i in nc.variables])) for j in [1,2,3,4]]
42 nt,nlay,nrow,ncol=nc.variables[V[3][0]].shape
43 nv=len(V[3])
44 nc.SDATE,nc.STIME=dt2jul(bdate)
45 nc.EDATE,nc.ETIME=dt2jul(edate)
46 nc.NOTE='grid Emission'
47 nc.NOTE=nc.NOTE+(60-len(nc.NOTE))*' '
48 #Name-names may encounter conflicts with newer versions of NCFs and PseudoNetCDFs.
49 #nc.NAME='EMISSIONS '
50 if 'ETFLAG' not in V[2]:
51 zz=nc.createVariable('ETFLAG',"i4",("TSTEP","VAR","DATE-TIME"))
52 if nt!=ntm:
53 for t in range(ntm):
54 sdate,stime=dt2jul(bdate+datetime.timedelta(days=t/24.))
55 nc.variables['TFLAG'][t,:,0]=[sdate for i in range(nv)]
56 nc.variables['TFLAG'][t,:,1]=[stime for i in range(nv)]
57 ndate,ntime=dt2jul(bdate+datetime.timedelta(days=(t+1)/24.))
58 nc.variables['ETFLAG'][t,:,0]=[ndate for i in range(nv)]
59 nc.variables['ETFLAG'][t,:,1]=[ntime for i in range(nv)]
60 for v in V[3]:
61 nc.variables[v][:]=0.
62 sdatetime=[jul2dt(nc.variables['TFLAG'][t,0,:]) for t in range(ntm)]
63
64
- 計算VOCs成分之準備
65 col=['PM','PM25', 'CO', 'NOX', 'NMHC', 'SOX'] #1NO 2NO2 3SO2 4NH3 5CCRS 6FCRS 7PAR
66 cole='tnmhc iso mono onmhc mbo'.split()
67 #define the crustals/primary sources
68 colv='OLE PAR TOL XYL FORM ALD2 ETH ISOP NR ETHA MEOH ETOH IOLE TERP ALDX PRPA BENZ ETHY ACET KET'.split()
69 icolv=[0,1,5,7]
70 NC=len(icolv)
71 c2v={colv[i]:colv[i] for i in icolv}
72 #name saroa num cas mw ole par tol xyl form ald2 eth isop nr
73 #ISOPRENE 43243 88 78795 68.12 1.0
74 #TERPENES 43123 19 136.23 0.5 6.0 1.5
75 #A-PINENE 43256 96 80568 136.24 0.5 6.0 1.5
76 #unidentified 140.0 0.5 8.5 0.5
77 #B-PINENE 43257 97 127913 136.24
78
79 #NSPEC=4;NCBM=5)
80 CBM=np.zeros(shape=(5,6))
81 CBM[1,4]=1.0
82 CBM[2,1:4]=[0.5 ,6.0 ,1.5]
83 CBM[3,1:4]=[0.5 ,6.0 ,1.5]
84 CBM[4,1:3]=[0.5 ,8.5]
85 MW=[float(i) for i in '0,68.0,136.24,136.23,140.0'.split(',')]
- 準備日變化係數檔案
86 #time variations for CBM's
87 #fact.csv is generated by BIO_FACT.f
88 dft=read_csv('fact.csv')
89 col=[i.replace(' ','') for i in dft.columns]
90 dft.columns=col
91 dft.ih=dft.ih-1
92 f24=np.zeros(shape=(24,NC))
93 for t in range(24):
94 boo=(dft.ih==t) & (dft.LAND==5)
95 idx=dft.loc[boo].index
96 if len(idx)==0:continue
97 icbm=np.array(dft.loc[idx,'ICBM'])-1
98 fcbm=np.array(dft.loc[idx,'FACT'])
99 f24[t,icbm]=fcbm[:]
100 f24=f24*24 #portions converted to rates
101 fact=np.zeros(shape=(ntm,NC))
102 hr=np.array([i.hour for i in sdatetime])
103 for i in range(NC):
104 fact[:,i]=f24[hr,i]
網格化、VOCs劃分
- 按照光化模式規格進行網格化、篩選出本月排放量
105 #note the df is discarded
106 df=disc(df)
107 df=df.loc[df.mon==mo].reset_index(drop=True)
- VOCs劃分
108 NREC=len(df)
109 df['KgpY2gpHr']=[1000./24./monthrange(yr,mo)[1] for mo in df.mon]
110 OTVOC=df.onmhc+df.mbo
111 SPEC=np.zeros(shape=(NREC,5))
112 SPEC[:,1]=df.iso
113 SPEC[:,2]=OTVOC/2.
114 SPEC[:,3]=df.mono
115 SPEC[:,4]=OTVOC/2.
116 for isp in range(1,5):
117 SPEC[:,isp]=SPEC[:,isp]*df.KgpY2gpHr/MW[isp]
118 ssum=np.zeros(NREC)
119 for i in icolv: #NR not write
120 v=colv[i]
121 ii=icolv.index(i)
122 ssum=0.
123 for isp in range(1,4):
124 ssum+=SPEC[:,isp]*CBM[isp,ii]
125 df[v]=ssum
126
時間與空間之展開
127 #Expand to ntm*NREC
128 sdt,ix,iy=(np.zeros(shape=(ntm*NREC),dtype=int) for i in range(3))
129 val=np.zeros(shape=(ntm*NREC,NC))
130 idatetime=np.array([i for i in range(ntm)],dtype=int)
131 for t in range(ntm):
132 t1,t2=t*NREC,(t+1)*NREC
133 ix[t1:t2]=list(df.IX)
134 iy[t1:t2]=list(df.IY)
135 for t in range(ntm):
136 t1,t2=t*NREC,(t+1)*NREC
137 sdt[t1:t2]=idatetime[t]
138 for v in range(NC):
139 c=colv[icolv[v]]
140 val[t1:t2,v]=fact[t,v]*np.array(df[c])
141 dfT=DataFrame({'idt':sdt,'IX':ix,'IY':iy})
142 for v in range(NC):
143 c=colv[icolv[v]]
144 dfT[c]=val[:,v]
145
線性之DataFrame填入3維矩陣
- 每個污染項目逐一進行
146 #Filling to the template 147 df=dfT 148 for c in c2v: 149 if c not in df.columns:sys.exit() 150 if sum(df[c])==0.:continue 151 if c2v[c] not in V[3]:continue 152 #only positive values are filled 153 dfc=df.loc[df[c]>0] 154 #since the number of time schemes are limited, the filling process are done 155 #by each values as a batch. 156 #If one attempt to loop the df axis to the point of the record, it will be very slow to do so. 157 imn,jmn=min(dfc.IX),min(dfc.IY) 158 imx,jmx=max(max(dfc.IX)+abs(imn)+1,ncol), max(max(dfc.IY)+abs(jmn)+1,nrow) 159 if imn<0 and imx+imn<ncol:sys.exit('negative indexing error in i') 160 if jmn<0 and jmx+jmn<nrow:sys.exit('negative indexing error in j') 161 - 填入過程
- 注意nc檔案並不適用np.array的fancy indexing(line 169~171)
- 詳NC檔案多維度批次篩選
- 先使矩陣所有內容為0
- 3個相同長度的序列
(idt,iy,ix),分別為矩陣的3個標籤內容 - 第4個同長度的序列
ss,即為要給定的值 - 填入此序列
- 再整批匯入至
nc檔案 - 注意nc檔案並不適用np.array的fancy indexing(line 169~171)
- 詳NC檔案多維度批次篩選
162 z=np.zeros(shape=(ntm,jmx,imx))
163 idx=dfc.index
164 idt=np.array(dfc.loc[idx,'idt'])
165 iy=np.array(dfc.loc[idx,'IY'])
166 ix=np.array(dfc.loc[idx,'IX'])
167 ss=np.array(dfc.loc[idx,c])
168 #Note that negative indices are not bothersome and are only at the end of the axis.
169 z[idt,iy,ix]=ss
170 #also mapping whole matrix, NOT by parts
171 nc.variables[c2v[c]][:,0,:,:]=z[:,:nrow,:ncol]
172
173 #pncgen(nc, fname, format = 'uamiv')
174 nc.close()
結果檢核
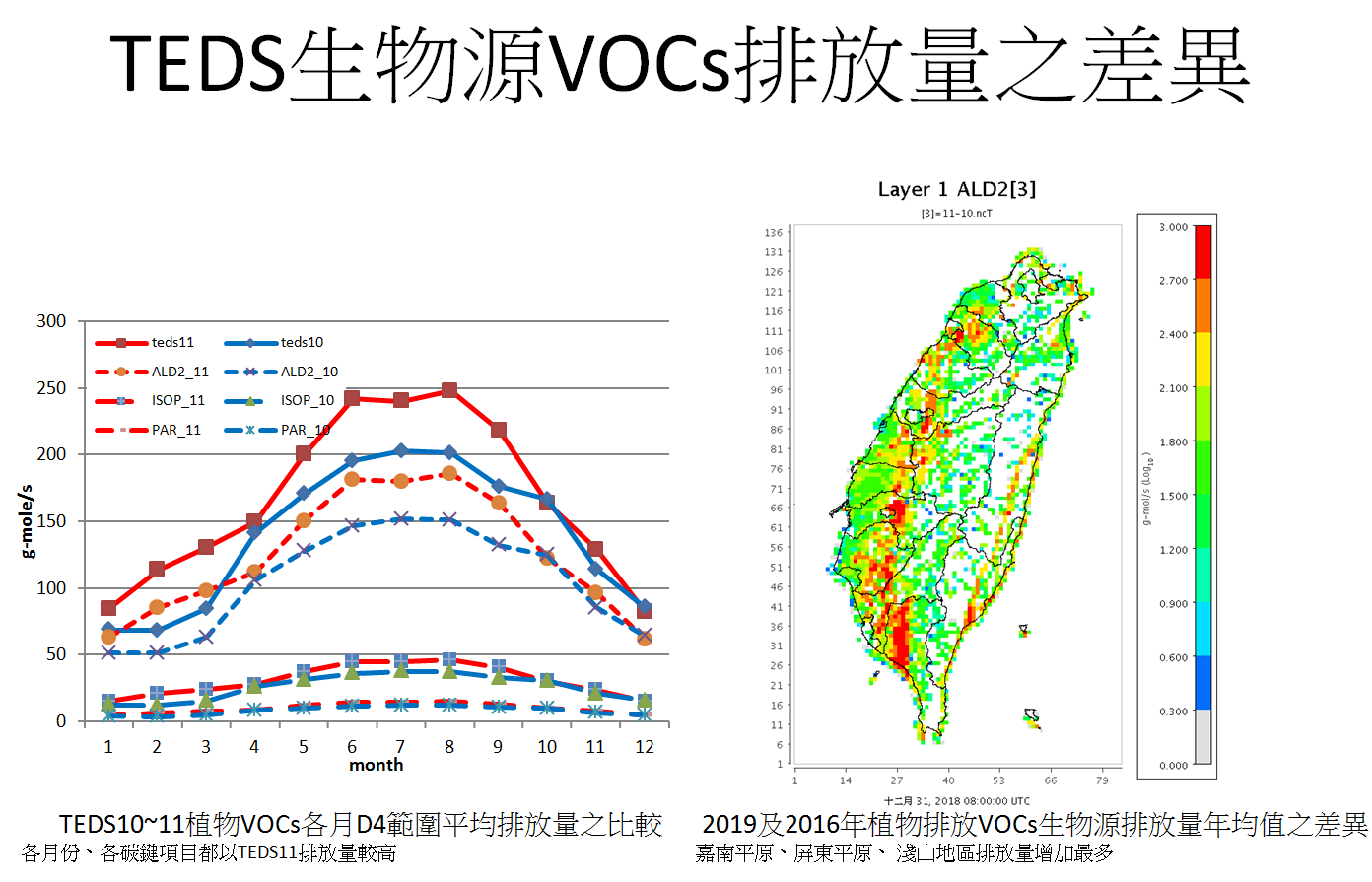
- 比較teds10及teds11之植物排放如圖
檔案下載
Download: python程式:bioginc.py